Medicine and Health
Design of 8-mer peptides that block *Clostridioides difficile* toxin A in intestinal cells
S. Sarma, C. M. Catella, et al.
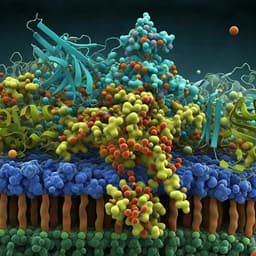
Discover groundbreaking research conducted by Sudeep Sarma, Carly M. Catella, and their team, focusing on peptide inhibitors that effectively block Toxin A of *Clostridioides difficile* in human colon epithelial cells. Their identified peptide, SA1, shows promising binding affinity, opening avenues for innovative treatments against a pressing global health issue.
~3 min • Beginner • English
Related Publications
Explore these studies to deepen your understanding of the subject.