Biology
Reconstructing aspects of human embryogenesis with pluripotent stem cells
B. Sozen, V. Jorgensen, et al.
The study addresses how to model and interrogate poorly understood mechanisms of early human embryogenesis, particularly pre- to peri-implantation events, given limited access to embryos and ethical restrictions. The authors hypothesize that human pluripotent stem cells grown under expanded pluripotency (EP) conditions (hEPSCs) can self-organize in 3D to recapitulate aspects of early embryonic lineage specification and morphogenesis, including blastocyst-like organization and early post-implantation remodeling. The work aims to establish a 3D culture platform from hEPSCs to capture polarization, cavitation, and initial segregation of epiblast (EPI), hypoblast (HYPO), and trophectoderm (TE)-like lineages, and to benchmark these structures morphologically and transcriptionally against natural human embryos.
Prior in vitro culture studies have enabled limited analysis of human embryos outside the body but are constrained by scarcity and ethics. Stem cell–derived embryo models in mouse and human have recapitulated select developmental stages, including mouse blastoids and human blastocyst-like structures from naïve or induced pluripotent cells. Expanded/extended pluripotency has been reported to confer broader lineage contributions, including extraembryonic potential in several species, motivating testing of hEPSCs for human embryo modeling. However, recent reports suggest EPSCs may reflect post-implantation epiblast-like states with restricted potency, and questions remain about TE versus amnion identity in human blastoids derived from iPSCs, highlighting potential decoupling between morphology and transcriptional identity.
- Cell conversion and maintenance: Human PSC lines (e.g., RUES2-GLR, ES1017) were converted to hEPSCs by ≥5 passages in hEP medium (DMEM/F12 + Neurobasal-A, N2, B27, GlutaMAX, NEAA, β-mercaptoethanol, pen/strep, 5% KSR) with LCDMYI supplements (LIF, CHIR99021, Dimethindene maleate, Minocycline, Y-27632), cultured on MEF feeders at 37 °C, 5% CO2, 20% O2. Mixed naïve/primed colony morphologies were observed.
- 3D aggregation: Using AggreWell microwells, 5–6 hEPSCs per microwell were seeded (hypoxia 5% O2). Culture medium: 50% human embryo IVF medium (CSCM-NXC), 25% hEP, 25% hTS base. To support survival and lineage emergence, the first 48 h included BMP4 (20 ng/mL), CHIR99021 (2 μM), FGF2 (40 ng/mL), Y-27632 (5 μM), and A83-01 (2 μM). After 48 h, A83-01 was removed and FGF2 reduced to 20 ng/mL. Low oxygen favored cavitation. Cavitated cysts emerged by day 3–4, with blastocyst-like morphology by day 6.
- Factor screening: Various growth factors/cytokines/small molecules were tested (including WNT3A 25–50 ng/mL) for effects on cavitation and lineage segregation.
- Polarity and lineage analyses: Immunostaining for PARD6, F-ACTIN, E-CADHERIN assessed apicobasal polarity during aggregation (22–72 h). GATA3 nuclear expression quantified relative to PARD6 apical enrichment. PLC pathway perturbations used U73122 (2 or 3 μM) and PLCB1 siRNA to test effects on polarity and GATA3 expression. Quantification used Fiji; statistical tests included Student’s t-test, Mann–Whitney, and Kruskal–Wallis with Dunn’s post hoc.
- Lineage marker assessment: Bulk qRT-PCR on pooled cystic structures profiled TE (PLAC8, CDX2, KRT8/18, GATA3), EPI (NANOG, POU5F1, KLF4), and HYPO (PDGFRA, GATA6, SOX17) genes. Immunofluorescence examined OCT4, SOX2, SOX17, FOXA2, KRT18, GATA3, E-CADHERIN localization in D4–D6 structures.
- Post-implantation-like culture (IVC): D5–D6 cystic structures were transferred to modified IVC1 medium (Advanced DMEM/F12 with FBS, GlutaMAX, pen/strep, ITS-X, β-estradiol, progesterone, N-acetyl-L-cysteine, IGF1, FGF2, FGF4, heparin) in ultra-low attachment plates for 24 h to assess peri-implantation remodeling (SOX2+ EPI-like lumen formation, outer extraembryonic-like markers, PODXL for lumen).
- scRNA-seq: Single cells from 2D hEPSCs, D5 cysts, D6 cysts after 24 h IVC, and natural D5/6 human blastocysts were captured with 10x Genomics (v3). MULTI-seq LMOs enabled multiplexing (except natural embryo). Analysis in Scanpy (normalization, HVG selection, clustering, UMAP). Lineage scoring used canonical gene sets (EPI, HYPO, TE). Gene program analysis used orthogonal NMF (PopAlign), hierarchical clustering, and Reactome-based enrichment (PDGF, Interleukin, VEGF, PI3K, STAT3, WNT).
- Co-assembly with human TSCs: After 24 h hEPSC aggregation, hTSCs were added in AggreWells (50% hEP/50% hTS medium) to test formation of cohesive TE-like epithelia and blastocyst-like architecture over ~4 days.
- Experimental design: Cavitated structures with clear lumen were selected for downstream analyses. No randomization/blinding; experiments typically repeated ≥2–3 times. Ethics approvals were obtained; culture terminated by day 8 in vitro.
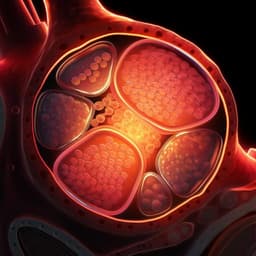
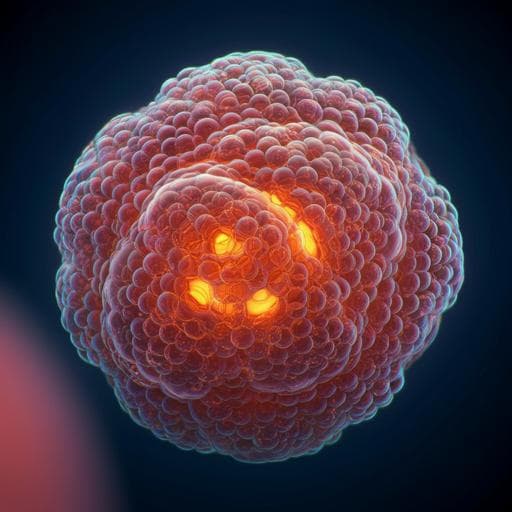
- Self-organization and morphology: Under optimized 3D conditions, hEPSCs formed cavitated cystic structures by day 3–4 and blastocyst-like morphology by day 6, with an outer epithelial layer, enlarged cavity, and acentric inner compartment. Average cell numbers and diameters were comparable to natural human blastocysts.
- Efficiency: By day 6, 7.2% of structures expressed markers consistent with three blastocyst lineages (SOX2 for EPI-like, SOX17/FOXA2 for HYPO-like, GATA3/KRT18 for TE-like) by immunofluorescence.
- Polarity and TE specification linkage: Early aggregates exhibited apical PARD6 and F-ACTIN enrichment with basolateral E-CADHERIN, indicative of polarization (≤48 h). GATA3 nuclear intensity was significantly higher in PARD6+ polarized cells versus PARD6− cells at day 2 (p=0.0033) and day 3 (p=0.0433). Pharmacologic PLC inhibition (U73122, 2–3 μM) reduced apical enrichment of PARD6 and F-ACTIN (p=0.0333) and diminished nuclear GATA3. PLCB1 siRNA knockdown decreased PARD6 apical enrichment at 48 h (p=0.006) and 72 h (p=0.0227) and reduced GATA3 expression, supporting a role for PLC-mediated polarity in promoting TE-associated GATA3.
- Lineage gene expression: qRT-PCR showed induction of TE-associated genes PLAC8, CDX2, KRT8, KRT18 upon cavitation; GATA3 increased only marginally. EPI factors NANOG and POU5F1 remained similar to 2D hEPSCs, while KLF4 was upregulated in cysts. HYPO determinants PDGFRA and GATA6 were enriched, whereas SOX17 did not mirror this trend.
- Protein localization: KRT18 enriched in outer layer; OCT4 and SOX17 in inner compartment (D5). SOX2 and FOXA2 co-marked inner compartment cells. At D4, some structures had outer GATA3 expression with retained OCT4. By D6, GATA3 persisted mainly in the cytoplasm (cytoplasmic 39.25%, nuclear 57.03%, none 3.70% across structures), and E-CADHERIN expression was poor, suggesting junctional deficiencies accompanying compromised TE-like identity.
- WNT3A effect: Exogenous WNT3A (25 or 50 ng/mL) did not significantly alter cavitation yield versus control at day 6 (ANOVA p=0.8473; n=450 control, n=483 25 ng/mL, n=419 50 ng/mL), contrasting with mouse reports.
- Post-implantation-like remodeling: Within 24 h in IVC, structures reorganized into peri-implantation-like morphology: 60.4% showed a SOX2+ inner compartment surrounded by KRT18/GATA3+ outer cells; FOXA2+ cells appeared in some structures (HYPO-like). SOX2+ cells became radially arranged around a central lumen confirmed by PODXL.
- scRNA-seq mapping: Cells clustered by sample identity. Lineage scoring identified EPI-like (ELC), HYPO-like (HLC), TE-like (TLC), and many undefined cells. hEP-structures had an overrepresentation of HLCs and relatively few TLCs compared to natural blastocysts, with many undefined states. A small GATA3+ subset in D5 clustered near natural TE but co-expressed amnion-associated markers (e.g., ISL1), suggesting amnion-like properties rather than bona fide TE in some TLCs. Over half of canonical lineage markers showed similar expression between ELCs and natural EPI (52/96 at D5; 50/96 at D6) and between HLCs and HYPO (52/96; 53/96). ECM genes (COL4A1, COL18A1, FN1) were aberrantly upregulated across multiple lineages. Reactome enrichment in hEP-structures highlighted PDGF, Interleukin, VEGF, PI3K, STAT3, and WNT signaling.
- Co-assembly with hTSCs: Adding human TSCs after 24 h hEPSC aggregation produced cystic structures with lineage marker expression (GATA3, SOX2, SOX17) but failed to form a cohesive TE-like epithelium and often generated multiple cavities, indicating the tested hTSC line could not rescue TE-like epithelium formation in this system.
The data support the hypothesis that hEPSCs can self-organize into 3D structures exhibiting several morphological and molecular features of early human embryogenesis, including polarization, cavitation, blastocyst-like organization, and limited post-implantation-like remodeling. However, the lineage composition and transcriptional identities are imperfect: HYPO-like cells are overrepresented, TE-like cells are underrepresented and often display amnion-like signatures, and many cells remain transcriptionally undefined. The findings underscore a decoupling between promising morphologies and accurate lineage-specific gene expression programs in stem cell-derived embryo models. Mechanistically, acquisition of apicobasal polarity via PLC signaling correlates with nuclear GATA3 and TE-like commitment, suggesting polarity-mediated cues influence extraembryonic lineage specification. Differences from mouse systems emerge (e.g., limited WNT3A effects), emphasizing species-specific regulation. Attempts to bolster TE using co-assembled human TSCs did not restore cohesive TE epithelium, possibly reflecting the hTSC state (villous cytotrophoblast-like) being more aligned with post-implantation. Overall, hEPSCs in this protocol likely occupy an intermediate, late epiblast-like state with restricted plasticity, limiting complete trans-differentiation into all blastocyst lineages.
This study establishes a 3D hEPSC-based platform that partially recapitulates early human embryogenesis, producing cavitated blastocyst-like structures that undergo limited peri-implantation-like reorganization. It links cell polarity (PLC-mediated) to TE-associated GATA3 expression and demonstrates selective activation of lineage programs. Single-cell analyses reveal substantial divergence from natural blastocysts, including overabundant HYPO-like states, scarce and potentially amnion-like TE-like cells, and widespread undefined states, evidencing a disconnect between morphology and transcriptional identity. Future work should refine hEPSC states and culture conditions to enhance TE specification and epithelial integrity (e.g., junctional maturation), disentangle amnion versus TE fates, optimize signaling environments (oxygen, ECM, growth factor dynamics), and develop or identify hTSC lines/states capable of pre-implantation-like co-assembly. Integrating multi-omic profiling and rigorous functional assays will be essential to advance faithful human embryo models.
- Low efficiency of full tri-lineage structures (7.2%) and disproportionate lineage representation (overabundant HYPO-like, scarce TE-like cells). Many cells remain transcriptionally undefined.
- TE-like identity is weak: marginal GATA3 upregulation, cytoplasmic localization at later times, poor E-CADHERIN at day 6, and amnion-like signatures in GATA3+ subsets.
- Aberrant ECM gene expression across lineages suggests non-physiological microenvironments and potential culture artifacts.
- WNT3A had no significant effect on cavitation, pointing to species-specific responses and/or suboptimal timing/dose regimens.
- Co-culture with available human TSCs failed to yield cohesive TE epithelium and produced multiple cavities; TSC state may be post-implantation-like.
- Selection bias: only cavitated structures with clear lumen were analyzed; non-cavitated aggregates were excluded.
- Experimental design limits: no predetermined sample sizes, no randomization or blinding; culture terminated by day ≤8 for ethical compliance, limiting assessment of later stages.
- Potential variability across hEPSC lines and passages with mixed naïve/primed features; outcomes may be line- and state-dependent.
Related Publications
Explore these studies to deepen your understanding of the subject.