Biology
Structure of the native pyruvate dehydrogenase complex reveals the mechanism of substrate insertion
J. Škerlová, J. Berndtsson, et al.
The pyruvate dehydrogenase complex (PDHc) is a megadalton multi-enzyme complex that converts pyruvate to acetyl-CoA, linking glycolysis to the citric acid cycle and lipid/sterol biosynthesis. It is crucial for glucose homeostasis and implicated in diseases such as obesity, type 2 diabetes, neurodegeneration, and cancer. PDHc comprises multiple copies of three enzymes: E1p (pyruvate dehydrogenase), E2p (dihydrolipoyl transacetylase), and E3 (dihydrolipoyl dehydrogenase). In mitochondria, an E3-binding protein and E1 kinases/phosphatases further regulate activity. E2 forms the structural and functional core, recruiting E1/E3 and mediating covalent substrate channeling via N-terminal lipoyl domains tethered by flexible linkers to a peripheral subunit-binding domain and a C-terminal catalytic domain. In Gram-negative bacteria, E2p catalytic domains assemble as eight tight trimers into a 24-meric cubic core with 432 symmetry. Each E. coli E2p bears three lipoyl domains; thus the 24-mer carries 72 lipoyl domains that deliver intermediates among active sites. Despite extensive structural studies, how lipoyl domains shuttle substrates into E2 active sites has remained unclear. The authors aimed to determine the native architecture and interactions of the E2p core and bound lipoyl domains in a resting state to elucidate the mechanism of substrate insertion.
- Sample source and state: Native PDHc isolated from E. coli K12 cells grown in M9 minimal medium with 0.5% succinate (no glucose), yielding low intracellular pyruvate and a resting-state complex.
- Purification: Cell lysis by sonication; ultracentrifugation; anti-FLAG affinity capture (cells carried a genomic FLAG on ubiF; purification outcome reproduced in wild type). Elution with 3xFLAG peptide; concentration to ~3 mg/ml; immediate cryo-EM grid prep or storage at −80 °C.
- Sample characterization: Silver-stained SDS-PAGE for purity; LC–MS/MS (QExactive Plus Orbitrap) to verify identity and intactness; western blot with anti-lipoate antibody to confirm lipoylation; mass spectrometry identified lipoylated residues K41, K144, K245 and assessed modification states.
- Enzyme activity assays: PDH and α-ketoglutarate dehydrogenase activities measured spectrophotometrically by NADH production at 340 nm in defined buffer conditions.
- Cryo-EM grid preparation and data collection: Glow-discharged Quantifoil grids; Vitrobot blotting at 4 °C, 100% humidity. Screening on Talos Arctica (200 kV, Falcon III). Data collected on Titan Krios (300 kV, K2 detector) across three datasets with varying grid types and protein concentrations; total 20,133 movies recorded.
- Image processing: cryoSPARC v2.15. Template picking yielded 799,790 particles; after 2D classifications, 42,266 particles advanced to 3D classification. Particles lacking ordered lipoyl domains (~30%) were excluded; final 29,434 particles refined with imposed octahedral symmetry (O) to 3.16 Å global resolution (0.143 FSC). Local resolution: catalytic core 2.8–3.5 Å; lipoyl domains 3.5–7.5 Å (0.5 FSC). Additional low-resolution 3D reconstruction of the whole PDHc (~4.5 MDa) without symmetry; outer shell components visualized in 2D classes after E2 core signal subtraction.
- Model building and validation: Docking of E. coli E2p catalytic domain (PDB 4n72) and innermost lipoyl domain (PDB 1qjo) using Phenix; manual model building in Coot; real-space refinements in Phenix/CCP-EM; validation with Phenix and MolProbity. Depositions: EMDB EMD-12104; PDB 7b9k.
- Mass spectrometry for lipoylation states: SP3 digestion; Exploris 480 LC–MS/MS; MaxQuant/Andromeda analysis defining oxidized, reduced, acetylated, and alkylated lipoylation states; localization probability >0.9.
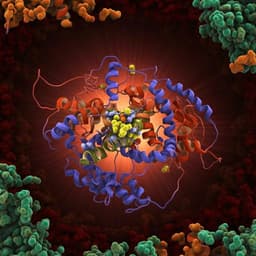
- Overall architecture: High-resolution cryo-EM structure of the native E. coli E2p 24-meric cubic core (octahedral symmetry) at 3.16 Å. The catalytic domains form trimers at cube vertices; each catalytic domain displays a bound lipoyl domain protruding as 24 visible "knobs". About 30% of particles lacked ordered lipoyl domains and were excluded.
- Resting-state composition: Cells grown without glucose produced low pyruvate; mass spectrometry showed lipoylation at K41, K144, K245 and only minor acetylated lipoyllysine, consistent with a resting state. Oxidation state (oxidized vs reduced dihydrolipoyllysine) could not be unambiguously assigned; map fit favored reduced dihydrolipoamide without S-acetyl.
- E2p core assembly details: Trimer–trimer interface at the 2-fold axis has ~840 Ų buried surface area with 24 residue pairs, dominated by hydrophobic interactions plus polar contacts (hydrophobic knob-into-hole of the short C-terminal helix). Additional polar stabilization (E447/Q443 with R627) likely reduces breathing motions seen in some species.
- Lipoyl domain binding mechanism: An extensive electrostatic interaction network guides docking. The lipoyl domain presents an acidic patch (including conserved E242, D244, and D213/217/218/220) engaging the positive dipole and R407 at the N-terminus of helix H1 of the catalytic domain; neighboring catalytic domain residues (e.g., K391', K396') may contribute transiently. Stable binding involves hydrogen bonds between LD loops (residues 213–215, 242–249) and catalytic loop (residues 536–544), plus van der Waals contact M248 (LD) with M543 (catalytic). Electronegative/positive surface patches are conserved across E2 families, supporting a general electrostatic navigation mechanism.
- Substrate insertion: The dihydrolipoyllysine (K245) is modeled deeply within the ~30 Å active-site channel at the interface of two catalytic domains. Its terminal sulfur (S8) is oriented toward the catalytic H603' and near S551 and D607'. A conserved D602'–R604' salt bridge stabilizes H603'. No CoA density was observed; comparisons with ligand-bound structures indicate an open channel in the absence of CoA, consistent with bacterial E2 lacking substrate-gated closure.
- Outer shell: Low-resolution reconstruction and 2D classes revealed peripheral subunit-binding domains and lipoyl domains associated with E1p/E3 in the outer shell, forming clusters near cube edges; due to flexibility and heterogeneity, individual E1/E3 components could not be fully modeled.
- Quantitative data: Final refinement used 29,434 particles; global resolution 3.16 Å; local resolution for catalytic core 2.8–3.5 Å and for lipoyl domains 3.5–7.5 Å.
The structure directly visualizes how a native, covalently lipoylated lipoyl domain binds the E2p catalytic domain and inserts the dihydrolipoyllysine into the E2 active-site channel, revealing the mechanism for substrate delivery during PDHc catalysis. The conserved electrostatic complementarity—an acidic patch on the lipoyl domain engaging the electropositive helix H1 and surrounding channel entrance—likely provides a general, nonspecific navigation system that accelerates productive encounters and positions the swinging arm for catalysis. The observed pose of dihydrolipoamide relative to H603', D607', and S551 aligns with known catalytic roles, situating the reactive sulfur near the general base histidine for the transacetylation step. The absence of CoA density and the open channel are consistent with bacterial E2p functioning without the CoA-triggered gating seen in mammalian E2b, suggesting evolutionary divergence in gating mechanisms (bacteria likely relying more on transcriptional regulation of operons). In the resting state with low pyruvate, the model rationalizes occupancy: oxidized lipoyl domains can bind E1/E3, reducing availability of E3 to reoxidize dihydrolipoamide and thereby stabilizing dihydrolipoamide bound at E2p active sites, as captured in the reconstruction. The trimer–trimer interface analysis supports conserved assembly principles (hydrophobic knob-into-hole) that allow different quaternary symmetries across species while ensuring a stable E2 core in E. coli.
This study provides a high-resolution cryo-EM structure of the native E. coli PDHc E2p cubic core in a resting state, revealing for the first time the molecular details of binding between an E2 catalytic domain and a native lipoyl domain with its dihydrolipoamide prosthetic group inserted into the active site. The work elucidates an electrostatics-driven navigation and docking mechanism for substrate insertion, maps conserved catalytic interactions within the active channel, and characterizes the conserved trimer–trimer interface underlying core assembly. These insights clarify how swinging lipoyl domains deliver intermediates within PDHc and likely generalize across E2-family systems. Future work could resolve the dynamic outer shell (E1/E3) at higher resolution, capture distinct catalytic states with substrates/products (e.g., CoA-bound, S-acetyldihydrolipoamide), determine lipoyl oxidation/acetylation states unambiguously in situ, and explore how variations in gating mechanisms correlate with evolutionary differences among bacteria and eukaryotes.
- The oxidation state of lipoyllysine (oxidized vs reduced) could not be unambiguously determined by mass spectrometry; the model favored reduced dihydrolipoamide without S-acetyl, but a mixture is likely.
- No CoA density was observed; the structure captures a resting state without substrates, not active catalytic transitions.
- Approximately 30% of particles lacked ordered lipoyl domains and were excluded; the remaining lipoyl domains exhibited lower local resolution (3.5–7.5 Å) than the catalytic core, limiting side-chain/detail assignment.
- The three E. coli lipoyl domains are highly similar; the map did not allow distinguishing which specific lipoyl domain was bound at each site.
- The outer shell (E1p/E3 and peripheral subunit-binding domains) was conformationally flexible and compositionally heterogeneous; low-resolution density precluded unambiguous assignment and complete modeling of these components.
Related Publications
Explore these studies to deepen your understanding of the subject.